INTRODUCTION
Schistosomiasis is a serious public health problem worldwide, particularly in Africa, South America, and Asia (Li et al. 2019). According to the World Health Organization (WHO), this disease is prevalent in over 78 countries, with approximately 779 million people at risk of infection (WHO 2022). In the Democratic Republic of Congo (DRC), schistosomiasis is endemic and constitutes a significant public health concern, particularly in the Kimpese region where prevalence can reach up to 68% (Mbuyi-Kalonji et al. 2020, Atila et al. 2021). The transmission in Africa involves freshwater snail species of the genera Bulinus and Biomphalaria (Zhang et al. 2022).
Historically, identifying these intermediate hosts relied on external morphological characteristics, including the shape, size and aperture of the shell, and to a lesser extent the internal characteristics of the gonads. However, this could sometimes lead to misidentifications because differences between closely related species can be very subtle and intraspecific variation in shell characteristics can be high (Adams 1861, Mandahl-Barth 1962). Advances in genomic sequencing techniques now allow for a more comprehensive species identification and understanding its diversity, including intraspecific diversity (Chevassus-au-Louis 2005). It should be noted that morphological identification is still very relevant, especially in low-income areas where molecular analyses do not have an easy access, but-an integrative approach combining morphological and molecular analysis is preferred. To gain a better understanding of the dynamics of the intermediate host snails of schistosomiasis, we conducted malacological surveys in the Kimpese region in DRC using this integrative approach.
Snails of the genus Amerianna Strand, 1928 (Gastropoda: Planorbidae) are endemic to the Australian mainland, Philippines and New Guinea. Most species have a sinistral, cylindrical carinate shell with many spiral ridges and non-punctate protoconch (Brown 1994, Ponder et al. 2023), thrive on wood and aquatic vegetation in ponds and tributaries, and feed on detritus and algae (Ponder et al. 2023). The current distribution and number of species of the genus Amerianna is poorly known (Ponder et al. 2023), which is probably linked with the taxonomic confusion among authors. For example, Brown (1983) reported a misidentification of Amerianna carinata collected from a lake in Ibadan, Nigeria in 1979 and 1983 and originally described as Bulinus rohlfsi Clessin, 1886, a synonym of Bulinus truncatus. In another instance, A. carinata was wrongly described as Bulinus indicus sp. nov. in India (Rao et al. 1994), raising concerns about schistosmiasis transmission in that area, but this was later correctly identified by Brown (1997). The confusion stems from the similar shell size and shape of both species, but unlike B. truncatus, A. carinata has sharply shouldered (angular) whorls, a non-punctate apex and extensive spiral and reticulate sculpture (Brown 1997). Amerianna carinata is not known as an intermediate host for Schistosoma spp., as several attempts by Walker to infect the species with Schistosoma haematobium and Schistosoma mansoni were unsuccessful (Walker 1988).
Jelnes & Fagbemi (1983) described an Amerianna sp. (possibly A. carinata) from the Botanical Garden and Agodi Garden in Ibadan Nigeria, and warned for the possible confusion with B. truncatus, which they collected from the same area. Their investigations showed that the enzyme profiles and chromosome numbers were clearly different, with 36 chromosomes for the former and 72 for the latter species. In the present paper, we report for the first time the presence of and molecular data on Amerianna sp. in the DRC, shedding light on the importance of complementing morphological identification with molecular techniques for targeted snail control.
MATERIAL AND METHODS
The survey was carried out in the Kimpese region in the western part of the DRC. Kimpese is one of the 30 health zones in the Kongo Central province, and is situated approximately 222 kilometres from the capital Kinshasa (Nkangu et al. 2021). The Kimpese health zone is particularly affected by schistosomiasis and was therefore selected for this study (Atila et al. 2021).
During July and October 2021, snails were collected in two sites of the Lukunga River, a right-bank tributary of the Congo River: in Wenze village (5°27′40.4″S, 14°21′42.5″E) and in Mbombo village (5°32′04.1″S, 14°22′02.1″E). Lukunga River was specifically targeted as the riverbanks are shallow and marshy. The river holds a significant importance as the primary tributary providing water to the surrounding communities (Droogmans 1901). Bulinus truncatus used in this study was sampled in the Tiki River in Kifua2 village (5°24′32.9″S, 14°21′10.7″E).
Snails were sampled up to five meters upstream and downstream of each site, according to De Clercq (1987). The collection was done through scooping with a circular sieve with two-millimeter mesh size mounted on a one-meter stick, complemented by manual picking. Sampling was done for 15 minutes at each site by two people (Bagalwa et al. 1996). Snail specimens were morphologically identified based on the characteristics such as shell opening and direction, shape, size, and coiling according to the identification keys of Mandahl-Barth (1962) and Brown (1980). The height, length, and width were measured using a graph paper.
Only snails morphologically identified as schistosome intermediate hosts were preserved in 70% ethanol and subsequently transported to the Royal Museum for Central Africa, Belgium, for further analysis and molecular identification. Before DNA extraction, the specimens were photographed according to the protocol described by Brecko et al. (2014). Upon closer examination based on these pictures, three specimens from two different sites showed identical morphology to each other, but slightly different from that of B. truncatus. These specimens were selected for sequencing together with a B. truncatus specimen from the Tiki River.
A Polymerase Chain Reaction (PCR) was conducted for the three snail samples. Snail DNA extraction was performed with the E.Z.N.A. Mollusc DNA Kit (OMEGA Bio-tek, USA) according to the manufacturer’s protocol. The DNA extracts were eluted in 75 microliter of elution buffer twice. Prior to PCR analysis, the samples were diluted 1:10. The PCR and sequencing were carried out following the protocols described by Kane & Rollison (1994) and Schols et al. (2019). A partial cytochrome c oxidase I (COI) fragment was amplified using the universal barcode primers (LCO1490-HCO2198) while a partial fragment of the internal transcribed spacer 1 region (ITS1 rDNA) was amplified using ETTS1 5’ TGCTTAAGTTCAGCGGGT 3’ and ETTS2 5’ TAACAAGGTTTCCGTAGGTGA 3’ primers from Kane & Rollinson (1994). The PCR reactions were done with the Simplex Taq DNA polymerase (Qiagen, Venlo, the Netherlands) following the respective protocols. The PCR products were purified for sequencing using the ExoSAP (Fermentas™) PCR purification protocol, and sequenced using forward and reverse primers and BigDye® chemistry (Macrogen™).
The resulting DNA sequences were checked for quality, cleaned, and assembled with Geneious (version 10.0.9). The sequences were compared with the GenBank and BOLD databases using the Basic Local Alignment Search Tool (BLAST) and the BOLD identification system. The phylogenetic analysis utilised the COI to obtain family-level information. Relevant reference Amerianna sequences from GenBank for the construction of the phylogenetic tree were selected, such as MT883680, MT88369 and MT883677 from Indo-Australia; MT883674 from Australia; MW386169 from India. The resulting selection was then aligned to our sequences with the muscle alignment algorithm in AliView (version 1.26) using default parameters. The best substitution model was determined before phylogenetic analysis through Modeltest implemented in MEGA version X (Tamura et al. 2021) and this was the General Time Reversible (GTR) model with gamma correction (with gamma 0.25). Bulinus truncatus sequences from our own study (HKT209), and from GenBank (MT707362 (Tanzania) and MG407341 (France)) were used as outgroups. DNA extracts from the three Amerianna snails are stored in the collection of the Royal Museum for Central Africa, RMCA.
The phylogenetic tree was constructed based on COI sequences using the Maximum Likelihood method and GTR + G model (Tamura et al. 2021).
RESULTS
The high-resolution stacking images are shown in Figs 1 and 2. We juxtapose images from Amerianna sp. (Fig. 1) with B. truncatus (Fig. 2) collected in a nearby site, two kilometres from the collection sites of Amerianna sp. Both species are sinistral and around 7 mm in size, but they slightly differ as the Amerianna shell has conspicuous whorls, it has a ridge or protrusion on the surface of the shell with a well-marked longitudinal shape which gives it a ribbed appearance. In contrast, the B. truncatus shell is smoother, with no visible protuberance, less pronounced whorls and less fragile compared to the Amerianna sp. shell. It is based on these differences and the genetic profile (see below) that the study specimen was assigned to Amerianna cf. carinata.
Fig. 1
High resolution image of Amerianna cf. carinata collected in the Lukunga River, DRC. The pictures were taken using a Canon EOS 600D camera fitted with a macro photo lens as described by Brecko et al. (2014). Scale bar 2 mm
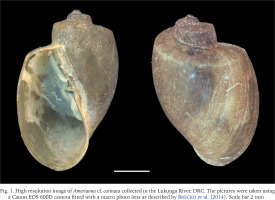
Fig. 2
High resolution image of a Bulinus truncatus specimen collected in the Tiki River in Kifua2 (5°24′32.9″S, 14°21′10.7″E), DRC. The pictures were taken using a Canon EOS 600D camera fitted with a macro photo lens as described by Brecko et al. (2014). Scale bar 2 mm

The ITS1 rDNA sequence was obtained for two specimens, and they were identical. The length of the longest generated consensus ITS1 rDNA sequence was 1,092 base pairs (bp), but the nucleotide BLAST query only generated matches for the ITS2 fragment. Based on this fragment, the similarity (98.74%) was the highest between our sequences and ITS2 of Amerianna sp. from Indo-Australian archipelago (MT893409), followed by Amerianna cf. leopoldi (95.98%; MT893405) and Amerianna cf. cumingi (93.05%; MT893404) generated in the same study from the same geographical area (Gauffre-Autelin et al. 2021).
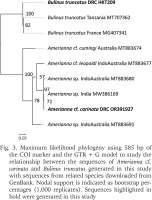
The length of the generated consensus COI sequence (obtained for one specimen) was 593 bp (GenBank accession number: OR391927). Our sequence was 100% identical to Amerianna sp. sequences from southern India (MW386168, MW386167, MW386166), very similar (99.15%) to Amerianna sp. from Indo-Australian archipelago (MT883680 and MT883679) and to a lesser extent to Amerianna cf. leopoldi (96%, MT883677). The phylogenetic tree constructed based on COI sequences is shown in Fig. 3.
Fig. 3
Maximum likelihood phylogeny using 585 bp of the COI marker and the GTR + G model to study the relationship between the sequences of Amerianna cf. carinata and Bulinus truncatus generated in this study with sequences from related species downloaded from GenBank. Nodal support is indicated as bootstrap percentages (1,000 replicates). Sequences highlighted in bold were generated in this study
DISCUSSION
The malacological, morphological and molecular investigations led to the discovery of an exotic snail species belonging to the genus Amerianna in DRC. The presence of this species in Central Africa has implications in snail distribution and ecology studies, possibly being falsely recorded as Bulinus snails. It is not easy to tell the difference between these two species by only taking into account the general morphological features such as the direction of the opening, the size or the shape of the shell. Consequently, Amerianna spp. can quickly go unnoticed when examining a mixed sample of snails with the naked eye in areas where they are not known to occur (Brown 1983).
Snail species of the Amerianna genus are endemic to Australia, but they were for the first time encountered outside their native range in Java in 1951, followed by Thailand, Nigeria, the Lesser Antilles, and in Southern India (Brown 1983, 1994, Walker 1988, Pointier et al. 2005, Gauffre-Autelin et al. 2021).
It is known that some newly introduced species can be invasive and compete with the local species (Appleton & Brackenbury 1997) in instances where the species’ reproduction and further dispersal are successful. The introduction of a new snail species in an environment can also have positive consequences if it outcompetes endemic snail species involved in disease transmission. For example, Biomphalaria glabrata, which is an intermediate host of S. mansoni, declined in abundance after the introduction of alien Marisa cornuarietis and Melanoides tuberculata in Guadeloupe (Pointier & Augustin 1999). In this study, the Amerianna species have been found in two different sites, so it has been able to establish viable populations, but only in low numbers. There are no reports of spreading in the literature, and this species can therefore not be considered as a pest species. Pointier et al. (2005) link the limited spreading of A. carinata to specific habitat preferences like artificial ponds and small ditches along riverbanks.
In this study, the phylogenetic tree (Fig. 3) indicates that the Amerianna sp. discovered at Kimpese is genetically identical to the Amerianna sp. found in India (Brown 1983, Appleton & Brackenbury 1997). This latter species was not determined to species level, as the authors await thorough taxonomic revision of the entire Amerianna genus (Appleton & Brackenbury 1997). Even though we agree with the authors that care should be taken, we tentatively infer that, based on all the previous studies and data (Brown 1983, 1994, Gauffre-Autelin et al. 2021), the species from Kimpese could be A. carinata, and hence refer to it as Amerianna cf. carinata (OR391927).
It remains to be explained how this species got introduced in these two natural, riverine sites in Kimpese, which are slightly atypical compared to the artificial water bodies described in the previous studies. The Lukunga River is a fast-flowing river, which might explain the absence of B. truncatus, which prefers stagnant or slow-flowing water. As pointed out by Jayachandran et al. (1928) and Pointier & Augustin (1999) snail species can disperse over large distances by means of the international aquatic plant trade, and the subsequent establishment is facilitated by man-made interventions like irrigation channels and the creation of artificial waterbodies. Other dispersal mechanisms could include transport through the feathers of waterfowl or on the feet of migratory birds, amphibians, vegetation transported through the watercourse; and certain types of fish like perch, in which the small snails’ embryos could burrow in their digestive tract (Jayachandran et al. (1928), Brown 1983, Chang et al. 2009). Snails can also use the hulls of boats or canoes to support their development and movement (Delongueville & Scaillet 2007). This last possibility is excluded in our context, since the Lukunga River, the collection site, is not connected by a navigable part of the Congo River which could be a gateway.
It is also not clear how long this Amerianna species has been present in the Kimpese region. It could have been overlooked in the previous malacological surveys, due to its resemblance to Bulinus spp. that are endemic in the DRC. More fieldwork is needed to answer this and the above questions and determine how far Amerianna cf. carinata has spread in Kimpese and possibly beyond.

