INTRODUCTION
Anthropogenic introductions of invasive species have significantly impacted native ecosystems, economic activity, and human health (Lowe et al. 2000, Simberloff et al. 2005, Didham et al. 2007, Pyšek et al. 2020). Invasive slugs reduce the populations of native terrestrial molluscs by decreasing their reproductive rates and increasing mortality of native species, as well as increasing hybridisation among them (Rollo 1983, Hatteland et al. 2015, Reise et al. 2020). Slugs also cause economic and sanitary damage to their non-native areas by plant feeding (Herbert 2010, Rowson et al. 2014b, Le Gall & Tooker 2017) and parasite transmission (Hollingsworth et al. 2013). Thus, investigating their current distribution, activity patterns, and control methods are important to reduce their long-term risks (Galvis & Moreno 2017, Morii et al. 2018, Nurinsiyah & Hausdorf 2019, Vendetti et al. 2019).
Milax gagates (Draparnaud, 1801) is a slug species with a wide non-native distribution. This species is assumed to be native to the western Mediterranean region, while it has been recorded in many countries in Europe, North and South America, and Oceania (Lovett & Black 1920, Wiktor 1987, Herbert 2010, Thomas et al. 2010). This species has been confused with congeners, such as M. nigricans (Philippi, 1836), because of their similar appearance, but their genital characteristics proved to be useful in distinguishing the species from other Milacidae species (Wiktor 1987, Hutchinson & Reise 2013, Rowson et al. 2014b). Recently, these two species have been identified by DNA barcoding using mitochondrial cytochrome c oxidase subunit I (COI) fragments (Turóci et al. 2023).
The non-native distribution of M. gagates includes Japan (Wiktor 1987, Barker 1999, Herbert 2010). This species was reported from Tokyo (Yamaguchi & Habe 1955) and Kanagawa Prefecture (Keio UHNSG 1991) in eastern Japan, and has been listed as an alien species in Japan (Kuroda 1963, Azuma 1982, Minato 1988, Subcommittee ISISGWPM 2002, Chiba Prefectural Government 2020). Field records of M. gagates are limited to these two prefectures in Japan, and the species has only been found during phytosanitary inspections at airports in recent years (Matsumoto & Kurozumi 2004). However, proper identification and morphological descriptions were not provided by the two brief field records, and to our knowledge, neither anatomical nor genetic examinations have been attempted on Japanese Milax specimens. Therefore, the recent establishment status and identity of Milax species have not yet been clarified in Japan.
We recently rediscovered Milax slugs on a river bank in Tokyo, Japan, and conducted species identification and habitat surveys, aiming to reveal the true identity of these milacid slugs and to find out whether they are established in Japan or remained single introductions.
MATERIAL AND METHODS
FIELD SURVEY AND SPECIMEN PREPARATION
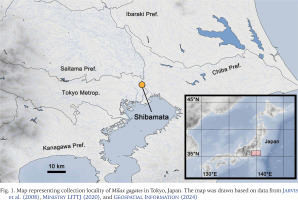
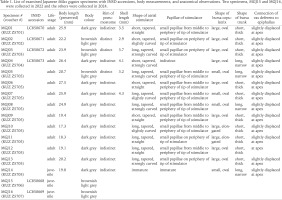
A field survey of slugs was conducted on the riverside of the Edogawa River in Shibamata, Katsushikaku, Tokyo Metropolitan, Japan (35°45'25"N, 139°52'54"E) on 14 May 2022, 17 April 2024, and 10 May 2024 (Fig. 1). The dominant vegetation of the habitat, rough abundance and habitat range of slugs, and sympatric terrestrial molluscs were visually investigated on 10 May 2024. The molluscs were identified following Azuma (1982), Minato (1989), Hwang et al. (2021), and MolluscaBase (2024). Among Milax slugs observed on the three survey days, two specimens collected on 14 May 2022 and 14 specimens collected on 10 May 2024, were used for morphological and/or genetic analyses in this study. They were stored at 13 °C in a refrigerator until further analysis. The specimens were prepared and photographed according to the method described by Turóci et al. (2023). After the morphological and genetic analyses, the specimens were deposited at the Zoological Collection of Kyoto University (KUZ Z5701–Z5707).
MORPHOLOGICAL EXAMINATION
The 14 slugs collected on 10 May 2024 were examined. The external morphology of the living and preserved specimens was photographed using a D7500 camera (Nikon Corporation, Tokyo, Japan) with a Nikon-compatible SP 90 mm f /2.8 1:1 macro lens (Tamron Co., Ltd., Saitama, Japan). The specimens were dissected using a YS05T stereomicroscope (Micronet Inc., Kawaguchi, Japan). The reproductive organs were photographed using a Swiftcam SC1003-CK camera (SwiftCam Technologies Group Co., Ltd., Kwai Chung, Hong Kong) attached to the stereomicroscope. Based on the morphological distinctiveness of Milax species indicated previously (Wiktor 1987, Hutchinson & Reise 2013, Turóci et al. 2023), we focused on the shapes of the atrial stimulator. In addition, position of the vas deferens outlet, shape and size of the bursa copulatrix, and body colour, which has been identified as less reliable (Hutchinson & Reise 2013, Turóci et al. 2023), was also investigated. After dissection, the shells and radulae were extracted from six and five specimens, respectively, using 1 M sodium hydroxide. According to Germain (1930), Wiktor (1987), and Rowson et al. (2014b), shell and radula morphology between the previous observations and the present specimens were compared. The body length of the preserved specimen and shell length were measured using ImageJ version 1.51 (Schneider et al. 2012). The extracted radulae were photographed using a JSM-6510LV scanning electron microscope (JEOL Ltd., Tokyo, Japan) after platinum coating using a JEC-3000FC (JEOL Ltd.).
GENETIC ANALYSIS
Morphological identification of the dissected specimens was confirmed using a partial region of mitochondrial COI. Genomic DNA was extracted from two specimens collected on 14 May 2022 and four specimens collected on 10 May 2024, following the procedure outlined by Okamoto et al. (2006). Polymerase chain reaction and nucleotide sequencing with the methods by Sawada et al. (2021) using the primers LCO1490 (5'-GGTCAACAAATCATAAAGATATTGG-3') and HCO2198 (5'-TAAACTTCAGGGTGACCAAAAAATCA-3') (Folmer et al. 1994). The newly obtained sequences were deposited in the International Nucleotide Sequence Database (INSD) through the DNA Databank of Japan (LC850668–LC850673). Genetic variations of the COI sequence were visually evaluated using MEGA version 11 (Tamura et al. 2021) against 20 sequences of M. gagates (including sequences labelled as Milax sp.) and M. nigricans provided by Rowson et al. (2014a), Gómez-Rodríguez et al. (2019), and Turóci et al. (2023). In addition, their haplotype relationships were inferred by statistical parsimony (Clement et al. 2000) using popart v1.7 (Leigh & Bryant 2015).
RESULTS
IDENTIFICATION
Slugs collected from the banks of the Edogawa River had light to dark brownish grey bodies with a distinct middorsal keel on the posterior mantle edge to the tip of the tale (Figs 2–7). The sole was mostly brownish light grey, and the dark grey pneumostome rim was conspicuous on the lighter-bodied specimens. Well-developed genitalia were observed in 13 specimens, whereas they were immature in one specimen (Table 1). In mature specimens, the atrial stimulator in the genitalia was short to long, tapered, and almost straight or strongly curved at the tip (Figs 8–11). Three to four small papillae were observed from the middle to the periphery of the tip of the stimulator except in one specimen which possessed a stimulator without distinct papillae (Table 1). Most specimens had an oval, large bursa copulatrix, a short, thick bursa trunk, and vas deferens connected slightly displaced at the apex of the epiphallus. They possessed a 2.9–4.3 mm oval, thin shell inside the anterior dorsal mantle (Fig. 12).
Figs 2–7
Dorsal, lateral, and ventral views of living specimens of Milax gagates collected from Tokyo, Japan: 2–4 – KUZ Z5701; 5–7 – KUZ Z5702. Slugs were stored at 13 °C in a refrigerator for three days from collection to being photographed

Figs 8–14
Reproductive organs (8–11), shell (12), and radula (13–14) of Milax gagates collected from Tokyo, Japan: 8 – entire genitalia, KUZ Z5701; 9 – genitalia around atrium, KUZ Z5702; 10–11 – atrial stimulator, KUZ Z5701 (10), Z5702 (11); 12 – KUZ Z5705; 13 – rachidian and lateral teeth, KUZ Z5702; 14 – lateral and marginal teeth, KUZ Z5702. Slugs were stored at 13 °C in a refrigerator for three to four days from collection to being dissected. Arrows indicate papillae on the atrial stimulator. Abbreviations: atg – atrial gland; ag – albumen gland; bc – bursa copulatrix; epi – epiphallus; hd – hermaphroditic duct; hg – hermaphroditic gland; ovi – oviduct; p – penis; so – spermoviduct; sti – atrial stimulator; vd – vas deferens. Scale bars 5 mm (8), 1 mm (9, 12), 0.5 mm (10, 11), 0.1 mm (13, 14)
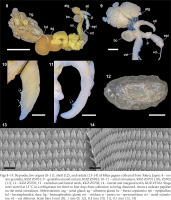
The radula possessed one rachidian and 32–44 lateral and marginal teeth on each side of the transverse row (Figs 13 & 14). The rachidian and lateral teeth had one arrow-headed large mesocone and one smaller ectocone and endocone on both sides. The mesocone was approximately 3.5 times longer than the ectocone and endocone in the inner rows of the radula, and these minor cusps were smaller in the outer rows. The marginal tooth possessed one pointed mesocone and was smaller in the outer rows.
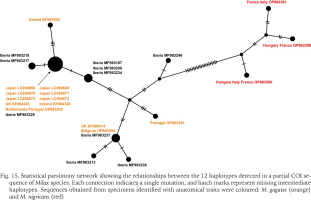
The 655 bp COI sequences obtained from six specimens showed the identical haplotype. These sequences matched completely in aligned positions with those of M. gagates from the UK (KF894243) and the Netherlands and Portugal (OP663302), and Milax sp. from the Iberian Peninsula (MF983226). The statistical parsimony network calculated based on the 12 haplotypes separated the sequences into two major groups by 21 mutational steps (Fig. 15). The network also showed that sequences obtained from the specimens of M. gagates (orange) and M. nigricans (red), which were identified with morphological traits, were divided at least in 26 mutational steps.
Fig. 15
Statistical parsimony network showing the relationships between the 12 haplotypes detected in a partial COI sequence of Milax species. Each connection indicates a single mutation, and hatch marks represent missing intermediate haplotypes. Sequences obtained from specimens identified with anatomical traits were coloured: M. gagates (orange) and M. nigricans (red)
HABITAT
Slugs were observed on paved roads and grassy areas at the same location on the banks of the Edogawa River on the three survey days. The distribution of the slugs was restricted to an area of approximately 0.3 ha and more than 150 Milax slugs with various body sizes were observed. The highest densities of slugs were found in sunny areas with short bushes, where herbaceous plants such as Avena fatua, Trifolium dubium, and Vicia sativa nigra were dominant. Milax slugs were observed sympatrically with the following five terrestrial mollusc species: Granulilimax fuscicornis Minato, 1989, Allopeas clavulinum kyotoense (Pilsbry et Hirase, 1904), Ambigolimax valentianus (Férussac, 1821), Acusta cf. sieboldtiana (Pfeiffer, 1850), Bradybaena similaris (Férussac, 1822).
DISCUSSION
In the present study, slugs collected from Japan were identified as M. gagates both morphologically and genetically. The anatomical characteristics observed in most Japanese specimens were consistent with those of M. gagates identified in previous studies (Wiktor 1987, Hutchinson & Reise 2013). Particularly, the papillae of the atrial stimulator were small or indistinct in all specimens, which is the reliable character for discriminating between M. gagates and M. nigricans (Hutchinson & Reise 2013). On the other hand, significant variations were observed in less diagnostic vas deferens, bursa copulatrix, bursa trunk, and body colour. The shell and radula morphology of Japanese slugs was similar to those of Milacidae species (Germain 1930, Wiktor 1987, Rowson et al. 2014b). The identical COI haplotype of the six Japanese slugs were the same as or close to those of M. gagates identified in previous studies (Rowson et al. 2014a, Turóci et al. 2023).
The haplotype network separated the 26 sequences into two major groups. The sequences of M. gagates and M. nigricans identified in the previous studies using anatomical traits belonged to different groups. The group of M. gagates included the sequences of M. gagates from UK, Ireland, Netherlands, Portugal, and Belgium (Rowson et al. 2014a, Turóci et al. 2023), Milax sp. from Iberian Peninsula (Gómez-Rodríguez et al. 2019), and the Japanese Milax obtained in this study. Furthermore, the haplotype of the Japanese specimens was identical to those from the UK, the Netherlands, Portugal, and the Iberian Peninsula. The native range of M. gagates has been estimated to be the coastal and insular regions of the western Mediterranean, and the species has artificially expanded into the regions where the same haplotypes as those in Japan have been obtained (Lovett & Black 1920, Wiktor 1987). Because all three sequences identical to Japanese ones were obtained outside the native range of M. gagates (Wiktor 1987), it is possible that the Japanese population was formed by secondary introduction from its non-native distribution, as well as direct transportation from its origin.
In the present study, M. gagates were found in the same locality in 2022 and 2024, and 150 mature and immature slugs were observed in 2024. Given that a twice-a-year reproduction has been revealed in this species (Wiktor 1987), it has likely established in this humid subtropical area in Japan.
Japanese Milax was observed with five terrestrial mollusc species. Among them, G. fuscicornis, Al. clavulinum kyotoense, and Ac. cf. sieboldtiana, are assumed native to Japan and Am. valentianus are thought to be non-native (Azuma 1982, Minato 1989, Hwang et al. 2021). There is still potential for both native and non-native status for B. similaris in Japan (Hirano et al. 2023). The introduction of alien slugs has contributed to the decline of native land snails through parasitic infections and predation (Hatteland et al. 2013, Howe et al. 2020). Feeding damage to crops and rare indigenous plants, and the spread of their diseases have also been partially attributed to non-native slugs, including M. gagates (Herbert 2010). Although the ecological impact of the Japanese M. gagates could not be evaluated in this study, they can contribute to the decline in the mollusc and plant species at the survey sites. Therefore, further investigation is required to evaluate its impact on native ecosystems and its expansion.